Microfluidics offers the possibility of running complex reactions on just a few square centimetres. Examples of current research are summarized below.
Medical University of Vienna: Method for sequencing millions of individual cells [1]
RNA sequencing is an important technology for researching cells and diseases. Single-cell sequencing in particular makes it possible to uncover the heterogeneity and diversity of our bodies. It is the central technology of the "Human Cell Atlas" for mapping all human cells. However, the method reaches its limits in very large projects, as it is time-consuming and very expensive. Scientists from the research group of Christoph Bock, at the CeMM Research Center for Molecular Medicine of the Austrian Academy of Sciences and professor at the Medical University of Vienna, have developed a new method for sequencing large numbers of individual cells more easily and cost-effectively [2].
Research into cells is an important basis for the development of personalized medicine. Five years ago, scientists around the world launched the "Human Cell Atlas" project with the aim of cataloging all cells in the human body. This data has helped, for example, to quickly identify those cell types that the coronavirus can infect particularly well.
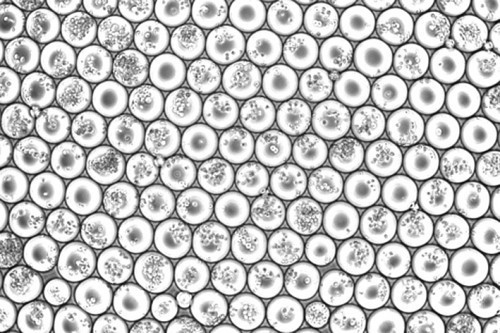 Emulsion droplets from a scifi-RNA-seq experiment loaded with a multiple of cells (Photo: Paul Datlinger, CeMM)In order to accelerate and improve the creation of such cell catalogs, Paul Datlinger and André F. Rendeiro from Christoph Bock's research group at CeMM developed a method to read out the activity of genes in a large number of individual cells simultaneously. This method, called "scifi-RNA-seq" (for "single-cell combinatorial fluidic indexing"), marks the RNA of many cells with barcodes in advance before the cells are dissolved in a microfluidic chip and their RNA is prepared for single-cell sequencing. These barcodes overcome a major problem of existing methods for single-cell sequencing.
Emulsion droplets from a scifi-RNA-seq experiment loaded with a multiple of cells (Photo: Paul Datlinger, CeMM)In order to accelerate and improve the creation of such cell catalogs, Paul Datlinger and André F. Rendeiro from Christoph Bock's research group at CeMM developed a method to read out the activity of genes in a large number of individual cells simultaneously. This method, called "scifi-RNA-seq" (for "single-cell combinatorial fluidic indexing"), marks the RNA of many cells with barcodes in advance before the cells are dissolved in a microfluidic chip and their RNA is prepared for single-cell sequencing. These barcodes overcome a major problem of existing methods for single-cell sequencing.
The method used to date faces the challenge that single-cell suspensions can only be loaded into the microfluidic chip at a very low concentration in order to avoid two cells ending up in the same emulsion droplet, which would result in a distorted cell profile. The majority of the emulsion droplets therefore had to remain empty in order to create a distance to the loaded droplets. The reagents were therefore only used very inefficiently.
By marking the cells upstream with various additional barcodes, the emulsion droplets in scifi-RNA-seq can be loaded with many cells at the same time and individual cells can still be analyzed. This saves time and costs. On the popular 10x genomics system, 15 times more single cells are measured with the new method. The additional barcodes also allow thousands of samples to be labeled and processed in a single microfluidic analysis. As part of the study, a CRISPR screen with single-cell sequencing was carried out in human T cells. In the future, the method should help to improve immunotherapies for the treatment of cancer, among other things.
The new method is particularly beneficial for projects that want to analyze a large number of cells or a large number of samples with single-cell sequencing. Scifi-RNA-seq enables efficient RNA sequencing of millions of individual cells and thus simplifies the characterization of complex tissues, organs and entire organisms. In the biomedical field, it is also often important to analyze a large number of single cells simultaneously, for example to discover rare stem cell populations in tumors or cancer cells in the blood. In addition, scifi-RNA-seq can help to ensure that drug screens and CRISPR screens are increasingly combined with high-resolution single-cell sequencing.
Fraunhofer IPM: New method for the detection of single molecules [3]
Resistance to antibiotics is constantly increasing worldwide. Researchers at the Fraunhofer Institute for Physical Measurement Techniques IPM, together with the LMU Munich, have developed a method to detect multi-resistant germs very quickly. The special feature: just a single DNA molecule is enough to detect the pathogen. In future, the platform is to be used in point-of-care diagnostics on hospital wards or in doctors' surgeries - as an alternative to the established PCR analysis or in combination with other diagnostic methods.
In the treatment of bacterial infections, the right antibiotic determines the success of the therapy. Selecting the right drug is particularly difficult when the disease is caused by multi-resistant pathogens that are insensitive to many antibiotics. The search for the most effective antibiotic often requires information about the bacterium's genome. However, this is usually not immediately available in the doctor's surgery, but only after a laboratory diagnosis. In order to speed up and simplify the process, Fraunhofer IPM is working with LMU Munich on the project (SiBoF), short for Signal Booster for Fluorescence Assays in Molecular Diagnostics, to develop a novel platform for pathogen detection using individual molecules on a microfluidic chip. The focus is on easy-to-use point-of-care (POC) detection. The project is funded by the Federal Ministry of Education and Research BMBF.
The portable, compact test platform has an automated fluidic system. All necessary reagents are stored upstream in the system. The injection-molded microfluidic chip is placed in a drawer in the test system, where it is supplied with the reagents by the fluidics before the optical analysis takes place. In the new method, part of the DNA strand of the pathogen is detected. A single DNA molecule that binds to a specific point on the microfluidic chip is sufficient for this. The chip contains fluidic channels whose surfaces have been prepared with binding sites for specific pathogens.
Typically, target DNA molecules are detected in vitro using specific fluorescent markers. The special feature of the new method developed by Fraunhofer IPM and LMU Munich: The researchers use antennas with nanometer-sized beads that amplify the optical signals of these markers. This eliminates the need for chemical amplification via the polymerase chain reaction (PCR). The optical antennas consist of nanometer-sized metal particles that bundle light in a tiny area and help to emit light - similar to macroscopic antennas with radio waves. These metal particles are chemically bonded to the surface of the chip.
A structure specially constructed by LMU Munich from DNA molecules, a so-called DNA origami, holds the two gold nanoparticles in place. Between these nanoparticles, the structure provides a binding site for the respective target molecule and a fluorescent marker. This patented design forms the basis for the novel assay technology. The particles, each 100 nanometers in size, serve as an antenna. In the hot spot between the two gold particles, a field amplification takes place due to plasmonic effects. If a fluorescent dye is placed there, the detectable longer-wave fluorescence radiation is amplified many times over. In this way, a single molecule can be detected with a small, compact optical device. Low concentrations of pathogens can be detected. The result is available after just one hour and is displayed on the monitor. This applies not only to multi-resistant germs, but also to any type of DNA molecule. In principle, the single-molecule assay can also be used for molecules other than DNA, such as RNA, antibodies/antigens or enzymes. The functionality of the method has been successfully confirmed by numerous tests.
At the heart of the POC device is a miniaturized high-resolution fluorescence microscope developed by Fraunhofer IPM. Special image analysis software identifies the individual molecules and thus enables the captured target molecules to be counted for a quantitative result. The fluorescence is stimulated by LEDs mounted underneath the cartridge with the fluidic channels. The patented system is available as a demonstrator. A module for sample preparation is currently still missing. The POC system for the specific detection of pathogens was presented at MEDICA 2021 in Düsseldorf.
Literature
[1] Medical University of Vienna
[2] Ultra-high-throughput single-cell RNA sequencing and perturbation screening with combinatorial fluidic indexing, Paul Datlinger, André F. Rendeiro, Thorina Boenke, Martin Senekowitsch, Thomas Krausgruber, Daniele Barreca, Christoph Bock; published on May 31, 2021 in Nature Methods, DOI: 10.1038/s41592-021-01153-z
[3] Fraunhofer IPM


