Using a new analytical method, Empa researchers have tracked viruses on their way through face masks and compared their failure on the filter layers of different mask types. The new method should now accelerate the development of surfaces that can kill viruses, as the team writes in the journal "Scientific Reports".
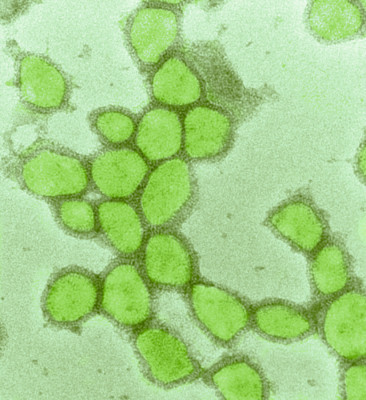 Fig. 2: Test viruses for the new method under the scanning electron microscope (recolored)The apparatus uses high pressure to chase the red-colored artificial saliva fluid with test particles through a stretched mask. In this way, the researchers simulate the process of droplet infection. The method established at Empa is currently being used by certified test centers to ensure the quality assurance of textile face masks, as a safe mask must meet demanding requirements:
Fig. 2: Test viruses for the new method under the scanning electron microscope (recolored)The apparatus uses high pressure to chase the red-colored artificial saliva fluid with test particles through a stretched mask. In this way, the researchers simulate the process of droplet infection. The method established at Empa is currently being used by certified test centers to ensure the quality assurance of textile face masks, as a safe mask must meet demanding requirements:
It must keep germs out, withstand splashing drops of saliva and at the same time allow breathing air to pass through.
The Empa researchers are now going one step further: "Images taken using a transmission electron microscope show that a few virus particles manage to find their way into the innermost layer of the mask close to the face. However, the images do not always reveal whether these viruses are still infectious," says Peter Wick from Empa's "Particles-Biology Interactions" laboratory in St. Gallen (Fig. 1). The researchers' goal: They want to find out at which point a virus fails at a multi-layer mask during a droplet infection, and which mask components should be more efficient. "This requires new analytical methods in order to precisely understand the protective function of newly developed technologies such as virus-killing coatings," says Empa researcher René Rossi from the "Biomimetic Membranes and Textiles" laboratory in St. Gallen.
This is precisely one of the goals of the "ReMask" project, in which research, industry and healthcare joined forces with Empa in the fight against the pandemic to develop new concepts for better, more comfortable and more sustainable face masks.
The new process is therefore based on the dye Rhodamine R18, which emits colored light. Non-hazardous, inactivated test viruses (Fig. 2) are used, which are coupled to R18 and thus become dying beauties: They light up in color as soon as they are damaged. "The fluorescence indicates reliably, quickly and cost-effectively when viruses have been killed," says Wick.
Based on the intensity with which a mask layer glows, the team was able to determine that most viruses in fabric and hygiene masks fail in the middle layer between the inner and outer layer of the mask. In FFP2 masks, the third of six layers glowed the most - here too, the central layer catches a particularly large number of viruses. The researchers recently published their results in the journal "Scientific Reports". These findings can now be used to optimize face masks.
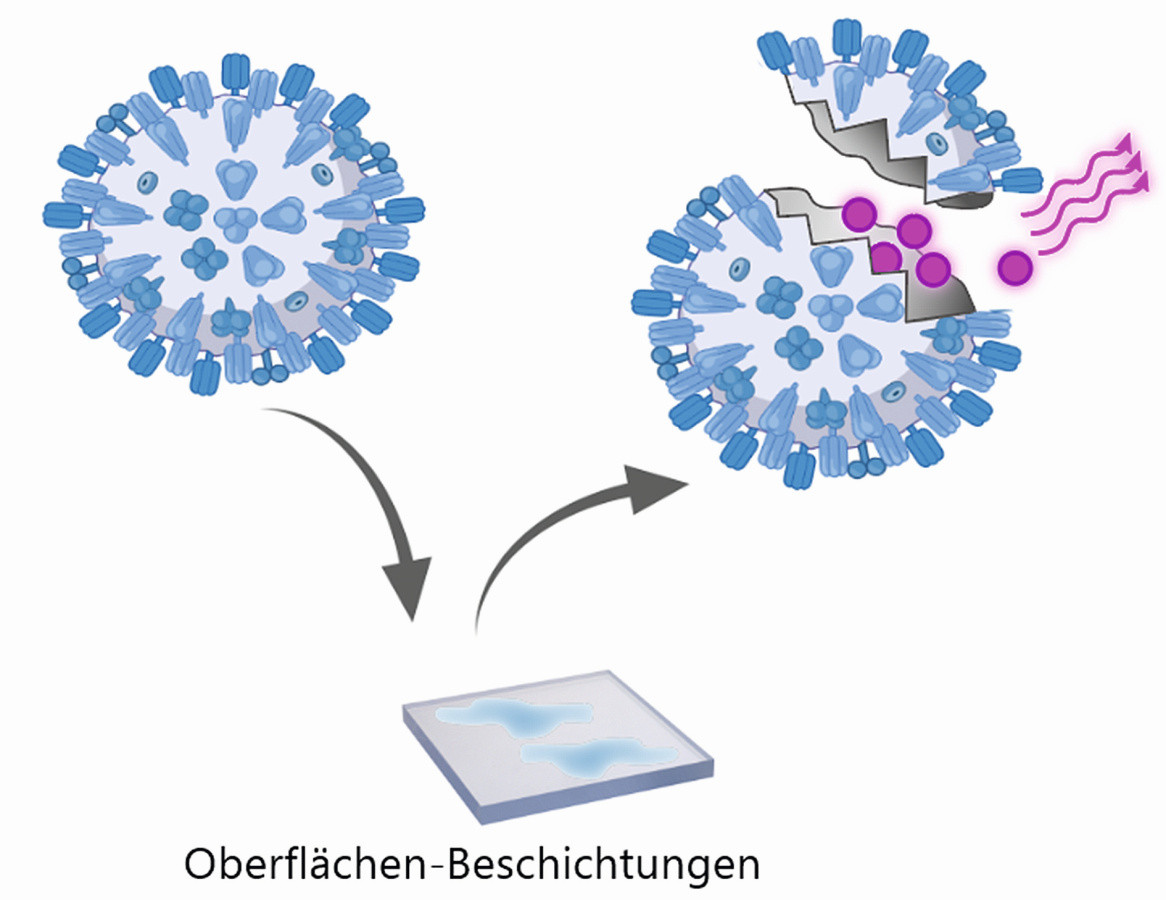
The new process can also accelerate the development of virus-killing surfaces (Fig. 3). "Surfaces with antiviral properties must comply with certain ISO standards, which entails complex standard tests," explains Wick. The fluorescence method used by the Empa researchers, on the other hand, could be a simpler, faster and more cost-effective addition to the current standards to determine whether a new type of coating can reliably kill viruses. This would be interesting both for smooth surfaces such as worktops or handles, as well as for coatings on textiles with a porous surface such as masks or filter systems. And with the new process, this knowledge could be integrated into the development process of technical and medical applications at a very early stage. According to Wick, this will speed up the introduction of new products, as only promising candidates will have to undergo the complex and cost-intensive standard tests.